Deep learning (DL) thrives on the availability of
large numbers of high quality images with reliable labels. Due
to the large size of whole slide images in digital pathology,
patches of manageable size are often mined for use in DL models.
These patches are often variable in quality, weakly supervised,
individually less informative, and noisily labelled. To improve
classification accuracy even with these noisy input and labels in
histopathology, we propose a novel method for robust feature
generation using an adversarial autoencoder (AAE) . We utilize
the likelihood of the features in the latent space of AAE as a
criterion to weigh the training samples. We propose different
weighing schemes for our framework and test our methods on two
publicly available histopathology datasets. We observe consistent
improvement in AUC scores using our methods, and conclude
that robust supervision strategies should be further explored for
computational pathology.
In summary, we address the problem of learning robust models for image classification in the face of label noise and weak supervision. We use adversarial autoencoders to get sample-wise weights for each training image. We assume that the samples that can deteriorate the model training will fall into the lesser likelihood regions of the class-specific distribution priors. This assumption eliminates the need for additional optimization steps to calculate the sample-wise weights. We also explore different schemes that can be used to weigh variants of cross entropy loss function for robust supervision.
Methodology
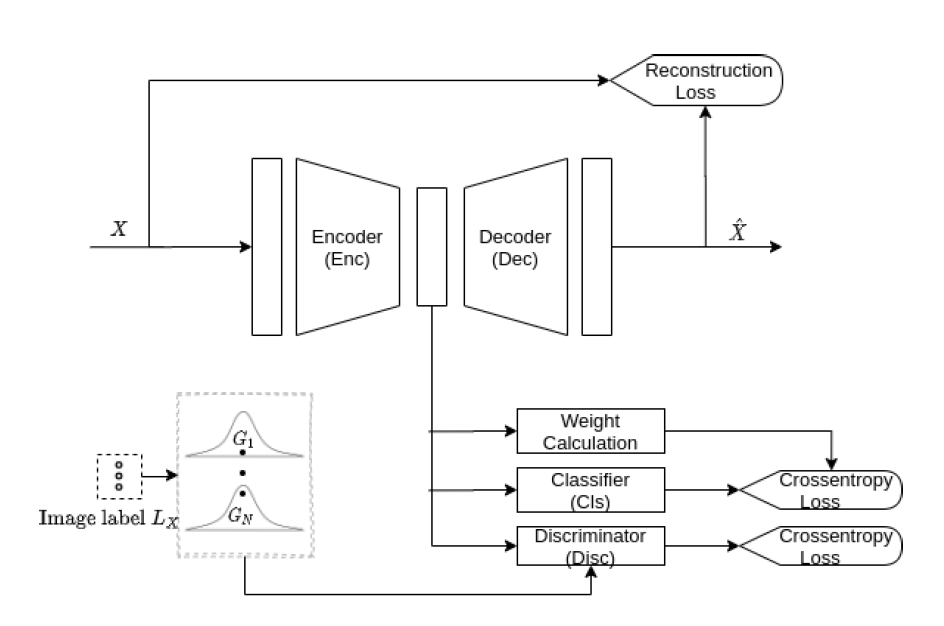
Our model has an encoder block that acts as a feature generator for the classifier and the AAE.
The discriminator block then performs adversarial training for the generated d-dimensional (d=32) latent features. The discriminator compares the encoded feature with a random vector sampled from its corresponding class specific d-dimensional Gaussian prior distribution. The encoder basically tries to generate samples that are optimized for the classifier while fooling the discriminator.
The decoder block ensures that all images belonging to a particular class are pulled towards a mean feature vector of its Gaussian prior.
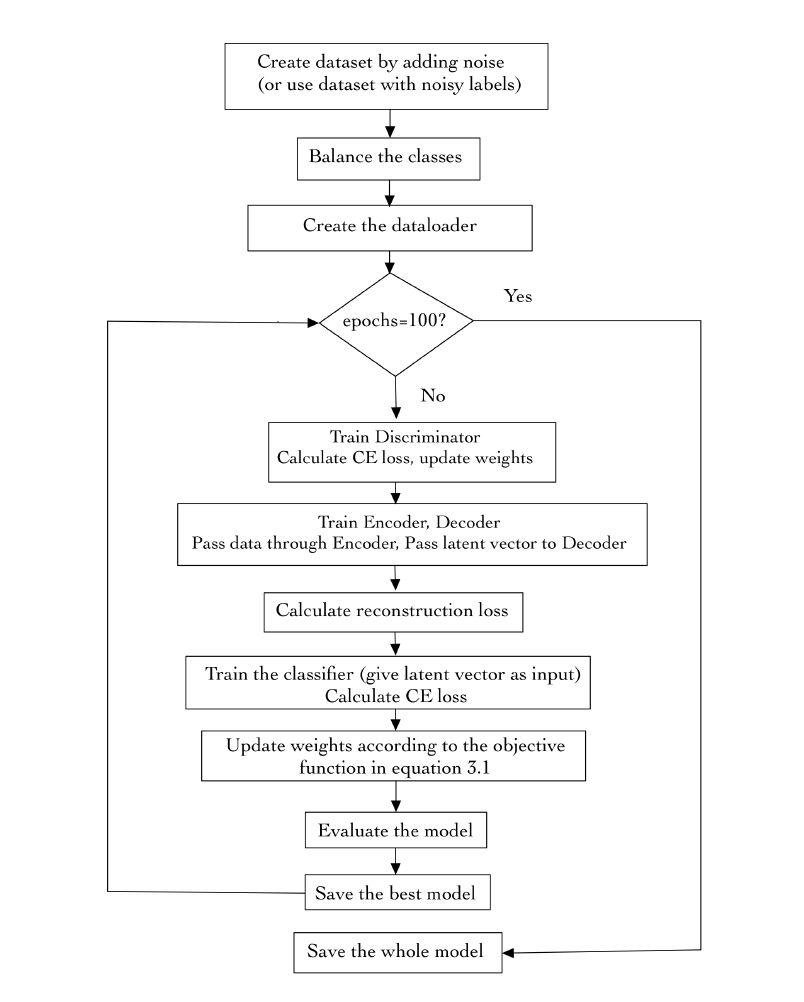
During the training
phase, feature generator and discriminator will perform the
following min-max game (equation 3.1) to generate the samples:
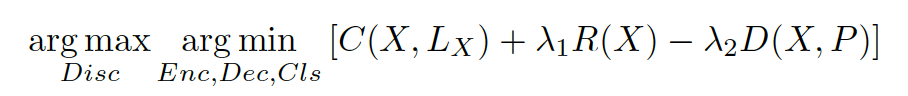
where C is the classification loss, R is the reconstruction loss,
and D is the discriminator loss. These losses are sample-wise
weighted cross-entropy, mean square error, and cross entropy
respectively in our scheme. Further, X is a training sample and
LX is its label, and P is a sample from the prior distribution.
When the discriminator reports low confidence in distinguishing a real Gaussian sample against the feature vector, the adversarial training is declared successful. Training the classifier separately on top of a well-trained AAE generator gave poor classifier performance because such a feature generator was agnostic to the classification task a priori.
Ablation studies
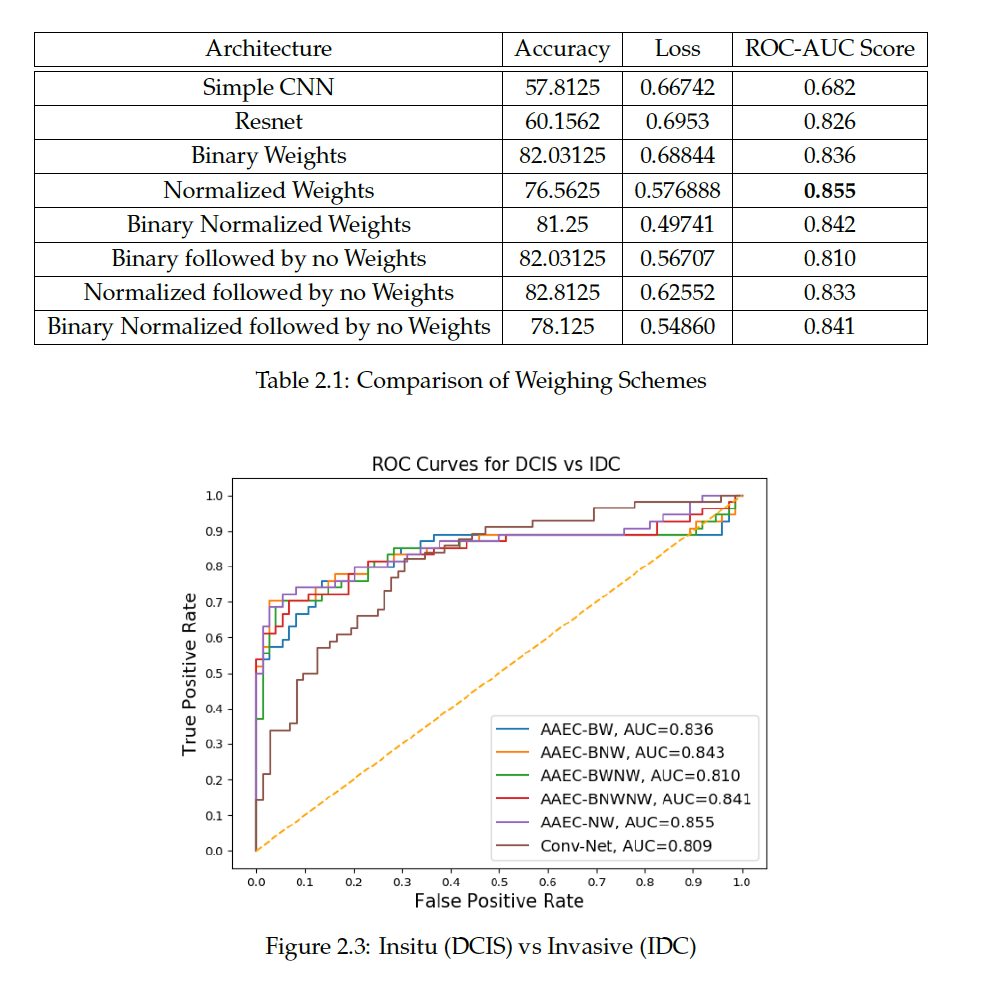
We explored the following sample-wise weighting schemes in the loss C to train the adversarial autoencoder based classifier (AAEC):
|
We further extended BW and BNW schemes, choosing the best of the models as the initial weights and continue training on these without any explicit weighting. We call these set of schemes that continue their training from BW and BNW without any weights as Binary Weighting-No Weighting (BWNW) scheme and Binary Normalised Weighting-No Weighting (BNWNW) scheme respectively.
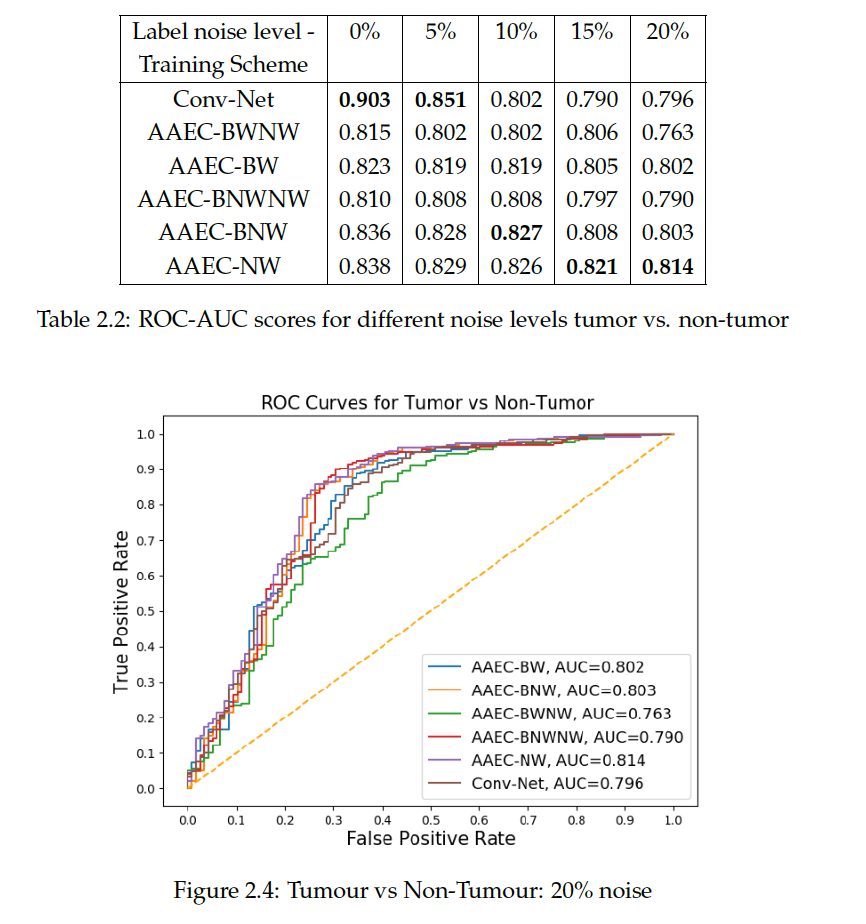
We evaluated the robustness of our models on clean held-out samples by manually curating the test cases before training the models. We show the AUC values and ROC curves on Breakhis and BACH dataset as shown below:
Conclusion and Future Work
From the results of the tumor versus non-tumor experiment, we observe that the performance of a regular CNN network worsens drastically with increasing noise. Further the DCIS versus IDC experiment shows that the robust weighting strategies perform much better than a conventional CNNs that implicitly overfits on noisy samples. The weighing strategies we found to be most robust in both the experiments. The experiments support the direction of further developing strategies for more robust histopathology, especially when the quality in image or strong supervision cannot be ensured. The next focus is to be on multi-class and unsupervised classification.